
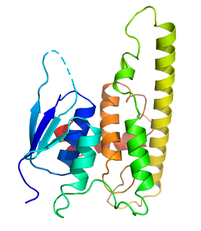
Mechanisms regulating Mitochondrial MorphologyMutations in the novel GST-like protein GDAP1 are responsible for subtypes of the neuropathy Charcot-Marie-Tooth disease (CMT). GDAP1 is localized to the outer mitochondria membrane where it plays a role in regulating the balance between mitochondrial fission and fusion. GDAP1 knockdown or the expression of some CMT mutants results in elongated and fused mitochondria while overexpression of wildtype GDAP1 results in fragmented mitochondria. The molecular mechanism(s) that drive these large scale cellular changes are not known. We are using biochemical, biophysical, structural and cell biology techniques to pinpoint regions of GDAP1 that are functionally important and to reveal GDAP1-mediated interactions that impact mitochondrial morphology. |
|
The role of PEBP1 in asthma-related lipid signaling |
|
|---|---|
|
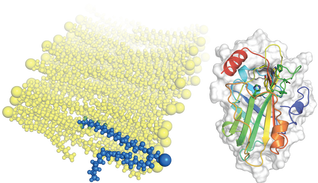
Despite the utility of anti-inflammatory and bronchodilating drugs, asthma often remains an episodic/fluctuating disease with severe episodes (or exacerbations) that contribute to thousands of deaths annually in the United States alone. We have information on the identity of the major protein players involved in asthma, specifically severe, exacerbation- prone asthma, but we lack information on the molecular mechanisms that govern their interactions or their consequences. We are working to understand the role of a critical adapter protein, PEBP1, in this process. PEBP1 has a relative plastic but highly conserved binding pocket. We are revealing how the binding pocket is utilized to bind lipids and how this impacts lipid signaling, which has dramatic cellular consequences. |
|
Revealing novel phage-host interactions |
|
|---|---|
|
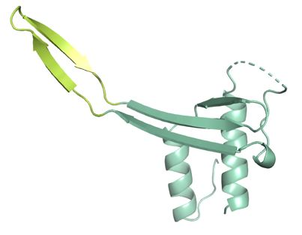
Mycobacteriophage are the most abundant entity on the planet and have been evolving with their mycobacterial hosts for billions of years. Metagenomic analysis of mycobacteriophages has revealed a common set of "structural" genes that encode for proteins that generate the phage assembly (capsid, tapemeasure, portal, etc), as well as others that regulate the generation and maintenance of the lysogen for temperature phages. Intensive efforts by many labs, including ours, has established the molecular basis that underlies the function of many of these proteins leading to many critical advances. There is however another class of phage proteins, which are typically organized on the other side of their linear genome, are often smaller, and which often have limited or no sequence homology to proteins of known structure or function. Many of these have been shown to mediate important phage-host interactions although the mechanisms are usually unknown. We are using structural, biochemical, and bioinformatics techniques to reveal structural information for novel phage proteins and combining this with molecular biology and functional data to identify new pathways by which phage interact and impact their bacterial hosts. |
|

 Research
Research